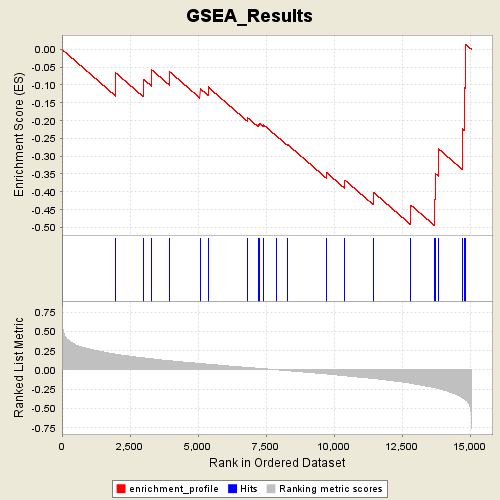
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Diabetes_collapsed.gct |
| Phenotype | DMT |
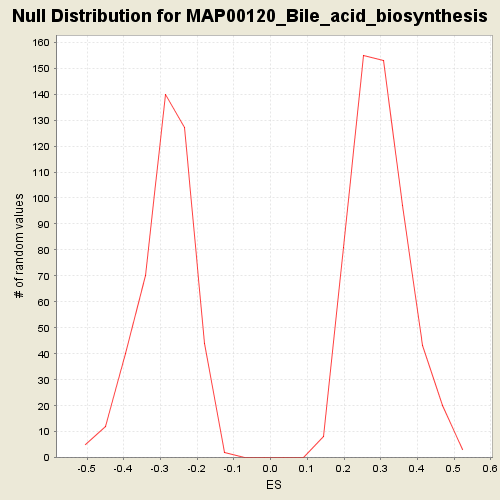
| GeneSet | C2.symbols.gmt#MAP00120_Bile_acid_biosynthesis |
| Enrichment Score (ES) | -0.49594736 |
| Normalized Enrichment Score (NES) | -1.7280037 |
| Nominal p-value | 0.006818182 |
| FDR q-value | 0.99574906 |
| FWER p-Value | 0.461 |
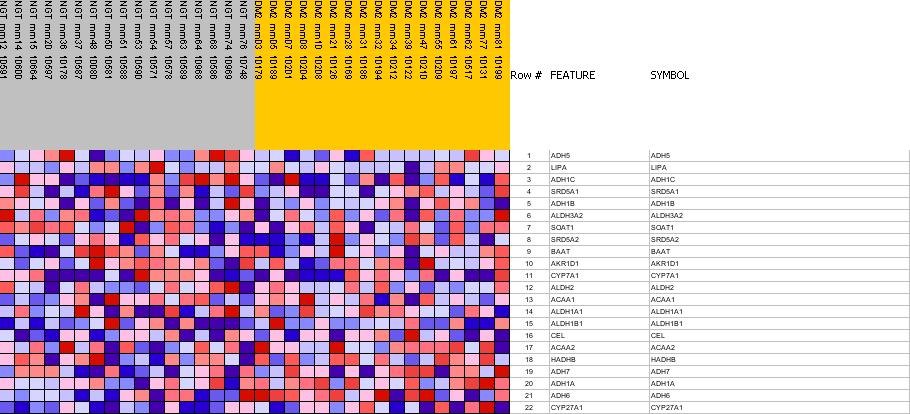
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RES | CORE_ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ADH5 | ADH5 StanfordSource, GeneCards | alcohol dehydrogenase 5 (class III), chi polypeptide | 1973 | 0.205 | -0.0657 | No |
| 2 | LIPA | LIPA StanfordSource, GeneCards | lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | 2997 | 0.157 | -0.0836 | No |
| 3 | ADH1C | ADH1C StanfordSource, GeneCards | alcohol dehydrogenase 1C (class I), gamma polypeptide | 3290 | 0.144 | -0.0568 | No |
| 4 | SRD5A1 | SRD5A1 StanfordSource, GeneCards | steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 3951 | 0.120 | -0.0623 | No |
| 5 | ADH1B | ADH1B StanfordSource, GeneCards | alcohol dehydrogenase IB (class I), beta polypeptide | 5074 | 0.084 | -0.1100 | No |
| 6 | ALDH3A2 | ALDH3A2 StanfordSource, GeneCards | aldehyde dehydrogenase 3 family, member A2 | 5369 | 0.075 | -0.1057 | No |
| 7 | SOAT1 | SOAT1 StanfordSource, GeneCards | sterol O-acyltransferase (acyl-Coenzyme A: cholesterol acyltransferase) 1 | 6821 | 0.033 | -0.1917 | No |
| 8 | SRD5A2 | SRD5A2 StanfordSource, GeneCards | steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2) | 7207 | 0.022 | -0.2104 | No |
| 9 | BAAT | BAAT StanfordSource, GeneCards | bile acid Coenzyme A: amino acid N-acyltransferase (glycine N-choloyltransferase) | 7263 | 0.020 | -0.2075 | No |
| 10 | AKR1D1 | AKR1D1 StanfordSource, GeneCards | aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) | 7409 | 0.016 | -0.2120 | No |
| 11 | CYP7A1 | CYP7A1 StanfordSource, GeneCards | cytochrome P450, family 7, subfamily A, polypeptide 1 | 7902 | 0.002 | -0.2440 | No |
| 12 | ALDH2 | ALDH2 StanfordSource, GeneCards | aldehyde dehydrogenase 2 family (mitochondrial) | 8292 | -0.009 | -0.2670 | No |
| 13 | ACAA1 | ACAA1 StanfordSource, GeneCards | acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | 9716 | -0.051 | -0.3454 | No |
| 14 | ALDH1A1 | ALDH1A1 StanfordSource, GeneCards | aldehyde dehydrogenase 1 family, member A1 | 10401 | -0.075 | -0.3671 | No |
| 15 | ALDH1B1 | ALDH1B1 StanfordSource, GeneCards | aldehyde dehydrogenase 1 family, member B1 | 11439 | -0.111 | -0.4007 | No |
| 16 | CEL | CEL StanfordSource, GeneCards | carboxyl ester lipase (bile salt-stimulated lipase) | 12827 | -0.174 | -0.4374 | No |
| 17 | ACAA2 | ACAA2 StanfordSource, GeneCards | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) | 13708 | -0.232 | -0.4219 | Yes |
| 18 | HADHB | HADHB StanfordSource, GeneCards | hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit | 13721 | -0.232 | -0.3484 | Yes |
| 19 | ADH7 | ADH7 StanfordSource, GeneCards | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 13832 | -0.241 | -0.2786 | Yes |
| 20 | ADH1A | ADH1A StanfordSource, GeneCards | alcohol dehydrogenase 1A (class I), alpha polypeptide | 14720 | -0.358 | -0.2229 | Yes |
| 21 | ADH6 | ADH6 StanfordSource, GeneCards | alcohol dehydrogenase 6 (class V) | 14792 | -0.377 | -0.1071 | Yes |
| 22 | CYP27A1 | CYP27A1 StanfordSource, GeneCards | cytochrome P450, family 27, subfamily A, polypeptide 1 | 14841 | -0.389 | 0.0142 | Yes |