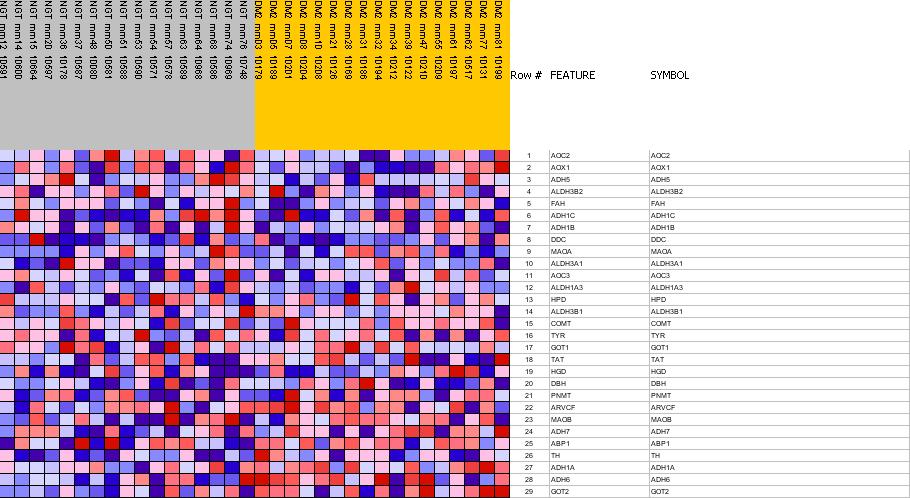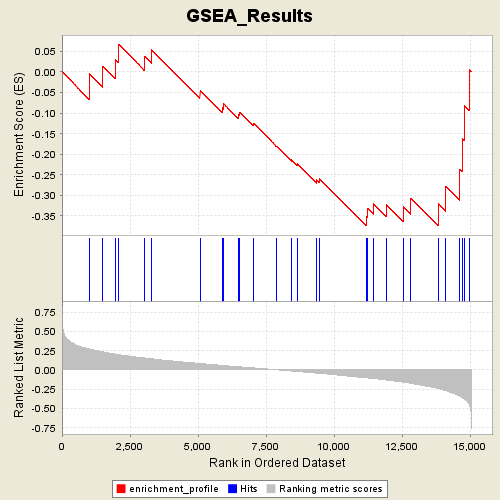
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Diabetes_collapsed.gct |
| Phenotype | DMT |
| GeneSet | C2.symbols.gmt#MAP00350_Tyrosine_metabolism |
| Enrichment Score (ES) | -0.3746339 |
| Normalized Enrichment Score (NES) | -1.3024019 |
| Nominal p-value | 0.10666667 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RES | CORE_ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | AOC2 | AOC2 StanfordSource, GeneCards | amine oxidase, copper containing 2 (retina-specific) | 999 | 0.273 | -0.0051 | No |
| 2 | AOX1 | AOX1 StanfordSource, GeneCards | aldehyde oxidase 1 | 1503 | 0.234 | 0.0141 | No |
| 3 | ADH5 | ADH5 StanfordSource, GeneCards | alcohol dehydrogenase 5 (class III), chi polypeptide | 1973 | 0.205 | 0.0291 | No |
| 4 | ALDH3B2 | ALDH3B2 StanfordSource, GeneCards | aldehyde dehydrogenase 3 family, member B2 | 2074 | 0.199 | 0.0671 | No |
| 5 | FAH | FAH StanfordSource, GeneCards | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 3037 | 0.155 | 0.0380 | No |
| 6 | ADH1C | ADH1C StanfordSource, GeneCards | alcohol dehydrogenase 1C (class I), gamma polypeptide | 3290 | 0.144 | 0.0538 | No |
| 7 | ADH1B | ADH1B StanfordSource, GeneCards | alcohol dehydrogenase IB (class I), beta polypeptide | 5074 | 0.084 | -0.0459 | No |
| 8 | DDC | DDC StanfordSource, GeneCards | dopa decarboxylase (aromatic L-amino acid decarboxylase) | 5886 | 0.060 | -0.0865 | No |
| 9 | MAOA | MAOA StanfordSource, GeneCards | monoamine oxidase A | 5951 | 0.057 | -0.0778 | No |
| 10 | ALDH3A1 | ALDH3A1 StanfordSource, GeneCards | aldehyde dehydrogenase 3 family, memberA1 | 6474 | 0.043 | -0.1030 | No |
| 11 | AOC3 | AOC3 StanfordSource, GeneCards | amine oxidase, copper containing 3 (vascular adhesion protein 1) | 6533 | 0.041 | -0.0976 | No |
| 12 | ALDH1A3 | ALDH1A3 StanfordSource, GeneCards | aldehyde dehydrogenase 1 family, member A3 | 7034 | 0.027 | -0.1248 | No |
| 13 | HPD | HPD StanfordSource, GeneCards | 4-hydroxyphenylpyruvate dioxygenase | 7882 | 0.003 | -0.1805 | No |
| 14 | ALDH3B1 | ALDH3B1 StanfordSource, GeneCards | aldehyde dehydrogenase 3 family, member B1 | 8429 | -0.013 | -0.2139 | No |
| 15 | COMT | COMT StanfordSource, GeneCards | catechol-O-methyltransferase | 8642 | -0.019 | -0.2238 | No |
| 16 | TYR | TYR StanfordSource, GeneCards | tyrosinase (oculocutaneous albinism IA) | 9350 | -0.040 | -0.2618 | No |
| 17 | GOT1 | GOT1 StanfordSource, GeneCards | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 9465 | -0.043 | -0.2596 | No |
| 18 | TAT | TAT StanfordSource, GeneCards | tyrosine aminotransferase | 11185 | -0.102 | -0.3511 | No |
| 19 | HGD | HGD StanfordSource, GeneCards | homogentisate 1,2-dioxygenase (homogentisate oxidase) | 11249 | -0.104 | -0.3319 | No |
| 20 | DBH | DBH StanfordSource, GeneCards | dopamine beta-hydroxylase (dopamine beta-monooxygenase) | 11456 | -0.111 | -0.3206 | No |
| 21 | PNMT | PNMT StanfordSource, GeneCards | phenylethanolamine N-methyltransferase | 11915 | -0.130 | -0.3219 | No |
| 22 | ARVCF | ARVCF StanfordSource, GeneCards | armadillo repeat gene deletes in velocardiofacial syndrome | 12544 | -0.158 | -0.3281 | No |
| 23 | MAOB | MAOB StanfordSource, GeneCards | monoamine oxidase B | 12825 | -0.173 | -0.3077 | No |
| 24 | ADH7 | ADH7 StanfordSource, GeneCards | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide | 13832 | -0.241 | -0.3204 | Yes |
| 25 | ABP1 | ABP1 StanfordSource, GeneCards | amiloride binding protein 1 (amine oxidase (copper-containing)) | 14114 | -0.268 | -0.2788 | Yes |
| 26 | TH | TH StanfordSource, GeneCards | tyrosine hydroxylase | 14615 | -0.337 | -0.2362 | Yes |
| 27 | ADH1A | ADH1A StanfordSource, GeneCards | alcohol dehydrogenase 1A (class I), alpha polypeptide | 14720 | -0.358 | -0.1625 | Yes |
| 28 | ADH6 | ADH6 StanfordSource, GeneCards | alcohol dehydrogenase 6 (class V) | 14792 | -0.377 | -0.0824 | Yes |
| 29 | GOT2 | GOT2 StanfordSource, GeneCards | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | 14968 | -0.444 | 0.0058 | Yes |