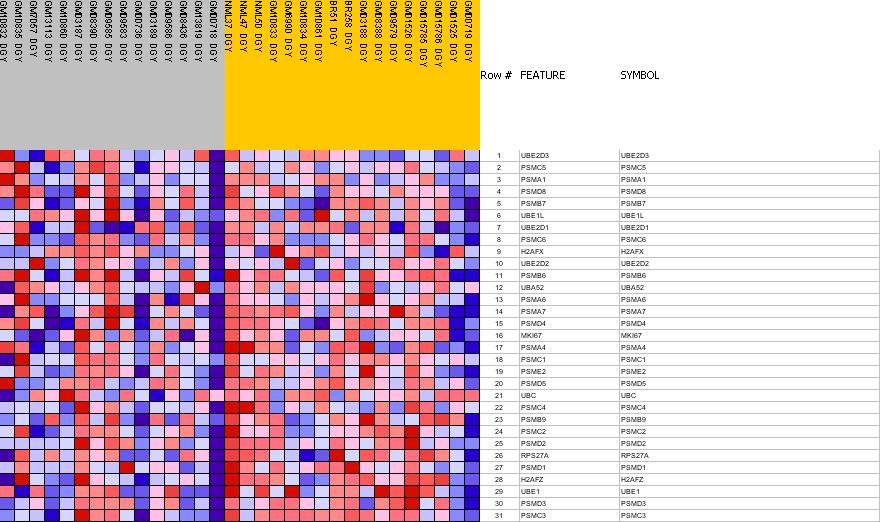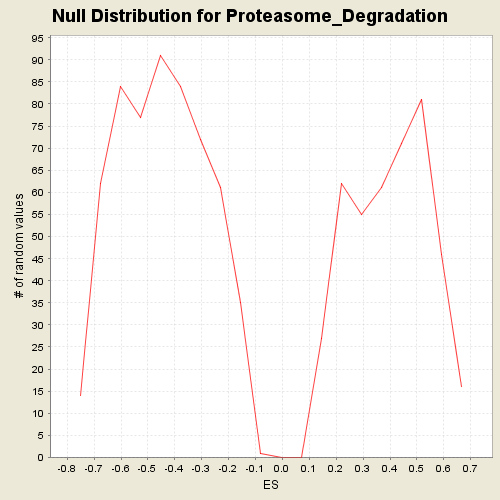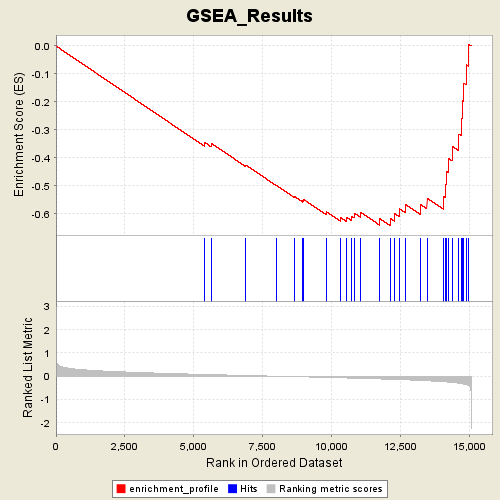
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Gender_collapsed.gct |
| Phenotype | FEMALE |
| GeneSet | C2.symbols.gmt#Proteasome_Degradation |
| Enrichment Score (ES) | -0.64275146 |
| Normalized Enrichment Score (NES) | -1.4459488 |
| Nominal p-value | 0.1222031 |
| FDR q-value | 0.85332966 |
| FWER p-Value | 0.804 |
| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RES | CORE_ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UBE2D3 | UBE2D3 StanfordSource, GeneCards | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | 5375 | 0.073 | -0.3435 | No |
| 2 | PSMC5 | PSMC5 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 5 | 5625 | 0.065 | -0.3473 | No |
| 3 | PSMA1 | PSMA1 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, alpha type, 1 | 6888 | 0.029 | -0.4256 | No |
| 4 | PSMD8 | PSMD8 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 8003 | -0.004 | -0.4990 | No |
| 5 | PSMB7 | PSMB7 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, beta type, 7 | 8655 | -0.022 | -0.5381 | No |
| 6 | UBE1L | UBE1L StanfordSource, GeneCards | ubiquitin-activating enzyme E1-like | 8944 | -0.029 | -0.5516 | No |
| 7 | UBE2D1 | UBE2D1 StanfordSource, GeneCards | ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) | 8980 | -0.030 | -0.5481 | No |
| 8 | PSMC6 | PSMC6 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 9812 | -0.055 | -0.5927 | No |
| 9 | H2AFX | H2AFX StanfordSource, GeneCards | H2A histone family, member X | 10308 | -0.070 | -0.6119 | No |
| 10 | UBE2D2 | UBE2D2 StanfordSource, GeneCards | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | 10518 | -0.077 | -0.6108 | No |
| 11 | PSMB6 | PSMB6 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, beta type, 6 | 10726 | -0.084 | -0.6082 | No |
| 12 | UBA52 | UBA52 StanfordSource, GeneCards | ubiquitin A-52 residue ribosomal protein fusion product 1 | 10820 | -0.087 | -0.5974 | No |
| 13 | PSMA6 | PSMA6 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, alpha type, 6 | 11032 | -0.094 | -0.5931 | No |
| 14 | PSMA7 | PSMA7 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, alpha type, 7 | 11744 | -0.119 | -0.6172 | No |
| 15 | PSMD4 | PSMD4 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 | 12129 | -0.134 | -0.6166 | Yes |
| 16 | MKI67 | MKI67 StanfordSource, GeneCards | antigen identified by monoclonal antibody Ki-67 | 12289 | -0.141 | -0.5998 | Yes |
| 17 | PSMA4 | PSMA4 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, alpha type, 4 | 12453 | -0.148 | -0.5819 | Yes |
| 18 | PSMC1 | PSMC1 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 1 | 12655 | -0.156 | -0.5648 | Yes |
| 19 | PSME2 | PSME2 StanfordSource, GeneCards | proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | 13221 | -0.184 | -0.5666 | Yes |
| 20 | PSMD5 | PSMD5 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 | 13452 | -0.198 | -0.5434 | Yes |
| 21 | UBC | UBC StanfordSource, GeneCards | ubiquitin C | 14050 | -0.240 | -0.5364 | Yes |
| 22 | PSMC4 | PSMC4 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 4 | 14137 | -0.247 | -0.4940 | Yes |
| 23 | PSMB9 | PSMB9 StanfordSource, GeneCards | proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional protease 2) | 14172 | -0.250 | -0.4477 | Yes |
| 24 | PSMC2 | PSMC2 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 14230 | -0.256 | -0.4017 | Yes |
| 25 | PSMD2 | PSMD2 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 14368 | -0.271 | -0.3580 | Yes |
| 26 | RPS27A | RPS27A StanfordSource, GeneCards | ribosomal protein S27a | 14610 | -0.304 | -0.3148 | Yes |
| 27 | PSMD1 | PSMD1 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 | 14698 | -0.323 | -0.2577 | Yes |
| 28 | H2AFZ | H2AFZ StanfordSource, GeneCards | H2A histone family, member Z | 14734 | -0.330 | -0.1957 | Yes |
| 29 | UBE1 | UBE1 StanfordSource, GeneCards | ubiquitin-activating enzyme E1 (A1S9T and BN75 temperature sensitivity complementing) | 14782 | -0.339 | -0.1328 | Yes |
| 30 | PSMD3 | PSMD3 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, non-ATPase, 3 | 14877 | -0.367 | -0.0676 | Yes |
| 31 | PSMC3 | PSMC3 StanfordSource, GeneCards | proteasome (prosome, macropain) 26S subunit, ATPase, 3 | 14966 | -0.408 | 0.0059 | Yes |